Note
Click here to download the full example code
Matching Image Keypoints by QAP Solvers
This example shows how to match image keypoints by graph matching solvers provided by pygmtools.
These solvers follow the Quadratic Assignment Problem formulation and can generally work out-of-box.
The matched images can be further processed for other downstream tasks.
# Author: Runzhong Wang <runzhong.wang@sjtu.edu.cn>
# Wenzheng Pan <pwz1121@sjtu.edu.cn>
#
# License: Mulan PSL v2 License
Note
The following solvers support QAP formulation, and are included in this example:
import jittor as jt # jittor backend
from jittor import Var, models # CV models
import pygmtools as pygm
import matplotlib.pyplot as plt # for plotting
from matplotlib.patches import ConnectionPatch # for plotting matching result
import scipy.io as sio # for loading .mat file
import scipy.spatial as spa # for Delaunay triangulation
from sklearn.decomposition import PCA as PCAdimReduc
import itertools
import numpy as np
from PIL import Image
pygm.BACKEND = 'jittor' # set default backend for pygmtools
# jt.flags.use_cuda = 1 # use cuda
Load the images
Images are from the Willow Object Class dataset (this dataset also available with the Benchmark of pygmtools,
see WillowObject).
The images are resized to 256x256.
obj_resize = (256, 256)
img1 = Image.open('../data/willow_duck_0001.png')
img2 = Image.open('../data/willow_duck_0002.png')
kpts1 = jt.Var(sio.loadmat('../data/willow_duck_0001.mat')['pts_coord'])
kpts2 = jt.Var(sio.loadmat('../data/willow_duck_0002.mat')['pts_coord'])
kpts1[0] = kpts1[0] * obj_resize[0] / img1.size[0]
kpts1[1] = kpts1[1] * obj_resize[1] / img1.size[1]
kpts2[0] = kpts2[0] * obj_resize[0] / img2.size[0]
kpts2[1] = kpts2[1] * obj_resize[1] / img2.size[1]
img1 = img1.resize(obj_resize, resample=Image.BILINEAR)
img2 = img2.resize(obj_resize, resample=Image.BILINEAR)
/mnt/c/Users/liber/OneDrive/Documents/2022/pygmtools/examples/jittor/plot_image_matching.py:62: DeprecationWarning: BILINEAR is deprecated and will be removed in Pillow 10 (2023-07-01). Use Resampling.BILINEAR instead.
img1 = img1.resize(obj_resize, resample=Image.BILINEAR)
/mnt/c/Users/liber/OneDrive/Documents/2022/pygmtools/examples/jittor/plot_image_matching.py:63: DeprecationWarning: BILINEAR is deprecated and will be removed in Pillow 10 (2023-07-01). Use Resampling.BILINEAR instead.
img2 = img2.resize(obj_resize, resample=Image.BILINEAR)
Visualize the images and keypoints
def plot_image_with_graph(img, kpt, A=None):
plt.imshow(img)
plt.scatter(kpt[0], kpt[1], c='w', edgecolors='k')
if A is not None:
for idx in jt.nonzero(A):
plt.plot((kpt[0, idx[0]], kpt[0, idx[1]]), (kpt[1, idx[0]], kpt[1, idx[1]]), 'k-')
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.title('Image 1')
plot_image_with_graph(img1, kpts1)
plt.subplot(1, 2, 2)
plt.title('Image 2')
plot_image_with_graph(img2, kpts2)

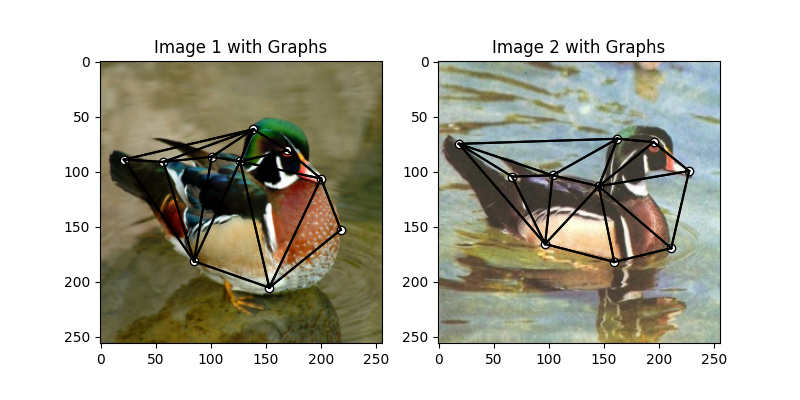
Build the graphs
Graph structures are built based on the geometric structure of the keypoint set. In this example, we refer to Delaunay triangulation.
def delaunay_triangulation(kpt):
d = spa.Delaunay(kpt.numpy().transpose())
A = jt.zeros((len(kpt[0]), len(kpt[0])))
for simplex in d.simplices:
for pair in itertools.permutations(simplex, 2):
A[pair] = 1
return A
A1 = delaunay_triangulation(kpts1)
A2 = delaunay_triangulation(kpts2)
We encode the length of edges as edge features
A1 = ((kpts1.unsqueeze(1) - kpts1.unsqueeze(2)) ** 2).sum(dim=0) * A1
A1 = (A1 / A1.max()).float32()
A2 = ((kpts2.unsqueeze(1) - kpts2.unsqueeze(2)) ** 2).sum(dim=0) * A2
A2 = (A2 / A2.max()).float32()
Visualize the graphs
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.title('Image 1 with Graphs')
plot_image_with_graph(img1, kpts1, A1)
plt.subplot(1, 2, 2)
plt.title('Image 2 with Graphs')
plot_image_with_graph(img2, kpts2, A2)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
Extract node features
Let’s adopt the VGG16 CNN model to extract node features.
vgg16_cnn = models.vgg16_bn(True)
jt_img1 = jt.Var(np.array(img1, dtype=np.float32) / 256).permute(2, 0, 1).unsqueeze(0) # shape: BxCxHxW
jt_img2 = jt.Var(np.array(img2, dtype=np.float32) / 256).permute(2, 0, 1).unsqueeze(0) # shape: BxCxHxW
with jt.no_grad():
feat1 = vgg16_cnn.features(jt_img1)
feat2 = vgg16_cnn.features(jt_img2)
Normalize the features
num_features = feat1.shape[1]
def local_response_norm(input: Var, size: int, alpha: float = 1e-4, beta: float = 0.75, k: float = 1.0) -> Var:
dim = input.ndim
if dim < 3:
raise ValueError(
"Expected 3D or higher dimensionality \
input (got {} dimensions)".format(
dim
)
)
if input.numel() == 0:
return input
div = input.multiply(input).unsqueeze(1)
if dim == 3:
div = jt.nn.pad(div, (0, 0, size // 2, (size - 1) // 2))
div = jt.nn.avg_pool2d(div, (size, 1), stride=1).squeeze(1)
else:
sizes = input.size()
div = div.view(sizes[0], 1, sizes[1], sizes[2], -1)
div = jt.nn.pad(div, (0, 0, 0, 0, size // 2, (size - 1) // 2))
div = jt.nn.AvgPool3d((size, 1, 1), stride=1)(div).squeeze(1)
div = div.view(sizes)
div = div.multiply(alpha).add(k).pow(beta)
return input / div
def l2norm(node_feat):
return local_response_norm(
node_feat, node_feat.shape[1] * 2, alpha=node_feat.shape[1] * 2, beta=0.5, k=0)
feat1 = l2norm(feat1)
feat2 = l2norm(feat2)
Up-sample the features to the original image size
feat1_upsample = jt.nn.interpolate(feat1, obj_resize, mode='bilinear')
feat2_upsample = jt.nn.interpolate(feat2, obj_resize, mode='bilinear')
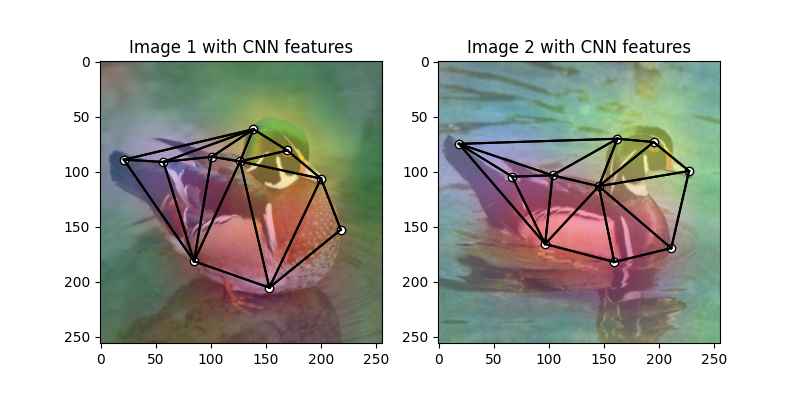
Visualize the extracted CNN feature (dimensionality reduction via principle component analysis)
pca_dim_reduc = PCAdimReduc(n_components=3, whiten=True)
feat_dim_reduc = pca_dim_reduc.fit_transform(
np.concatenate((
feat1_upsample.permute(0, 2, 3, 1).reshape(-1, num_features).numpy(),
feat2_upsample.permute(0, 2, 3, 1).reshape(-1, num_features).numpy()
), axis=0)
)
feat_dim_reduc = feat_dim_reduc / np.max(np.abs(feat_dim_reduc), axis=0, keepdims=True) / 2 + 0.5
feat1_dim_reduc = feat_dim_reduc[:obj_resize[0] * obj_resize[1], :]
feat2_dim_reduc = feat_dim_reduc[obj_resize[0] * obj_resize[1]:, :]
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.title('Image 1 with CNN features')
plot_image_with_graph(img1, kpts1, A1)
plt.imshow(feat1_dim_reduc.reshape(obj_resize[0], obj_resize[1], 3), alpha=0.5)
plt.subplot(1, 2, 2)
plt.title('Image 2 with CNN features')
plot_image_with_graph(img2, kpts2, A2)
plt.imshow(feat2_dim_reduc.reshape(obj_resize[0], obj_resize[1], 3), alpha=0.5)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
<matplotlib.image.AxesImage object at 0x7f4da498f280>
Extract node features by nearest interpolation
rounded_kpts1 = jt.round(kpts1).long()
rounded_kpts2 = jt.round(kpts2).long()
node1 = feat1_upsample[0, :, rounded_kpts1[0], rounded_kpts1[1]].t() # shape: NxC
node2 = feat2_upsample[0, :, rounded_kpts2[0], rounded_kpts2[1]].t() # shape: NxC
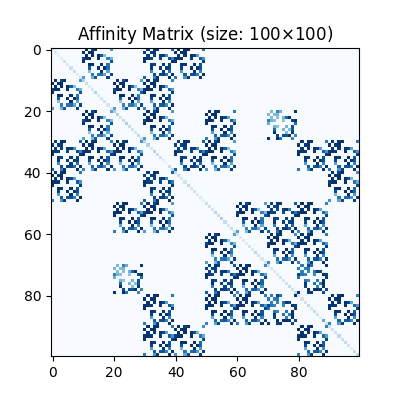
Build affinity matrix
We follow the formulation of Quadratic Assignment Problem (QAP):
where the first step is to build the affinity matrix (\(\mathbf{K}\))
conn1, edge1 = pygm.utils.dense_to_sparse(A1)
conn2, edge2 = pygm.utils.dense_to_sparse(A2)
import functools
gaussian_aff = functools.partial(pygm.utils.gaussian_aff_fn, sigma=1) # set affinity function
K = pygm.utils.build_aff_mat(node1, edge1, conn1, node2, edge2, conn2, edge_aff_fn=gaussian_aff)
Visualization of the affinity matrix. For graph matching problem with \(N\) nodes, the affinity matrix has \(N^2\times N^2\) elements because there are \(N^2\) edges in each graph.
Note
The diagonal elements are node affinities, the off-diagonal elements are edge features.
plt.figure(figsize=(4, 4))
plt.title(f'Affinity Matrix (size: {K.shape[0]}$\\times${K.shape[1]})')
plt.imshow(K.numpy(), cmap='Blues')
<matplotlib.image.AxesImage object at 0x7f4da4865430>
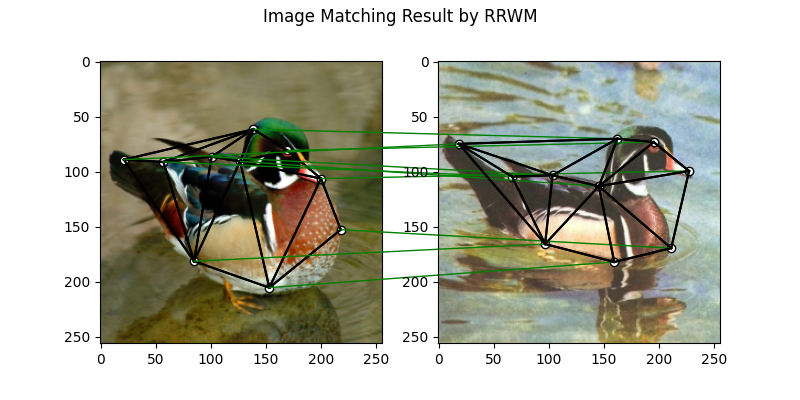
Solve graph matching problem by RRWM solver
See rrwm() for the API reference.
X = pygm.rrwm(K, kpts1.shape[1], kpts2.shape[1])
The output of RRWM is a soft matching matrix. Hungarian algorithm is then adopted to reach a discrete matching matrix
X = pygm.hungarian(X)
Plot the matching
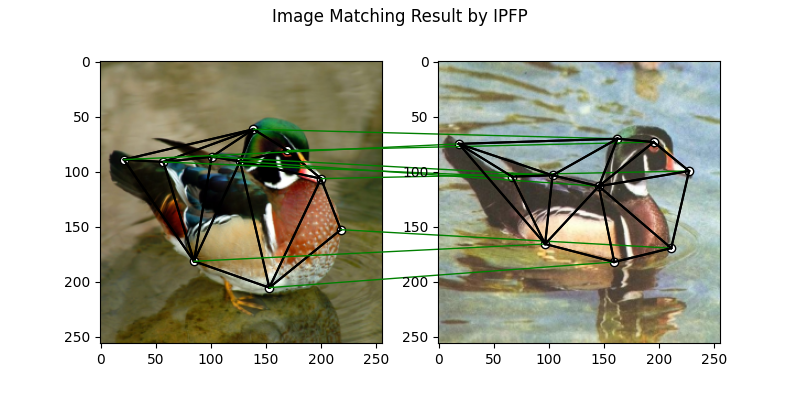
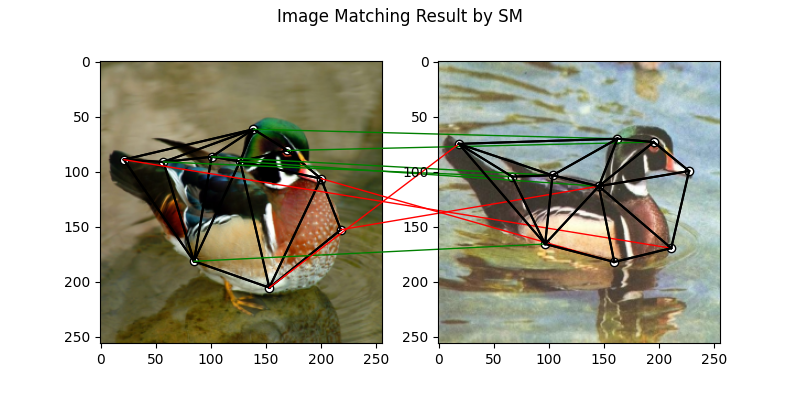
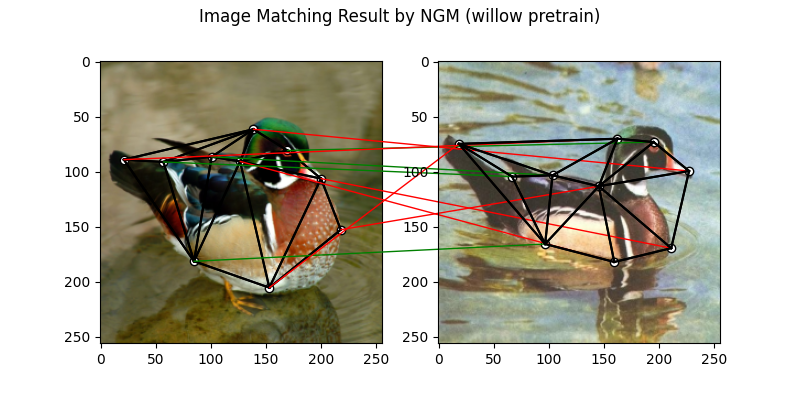
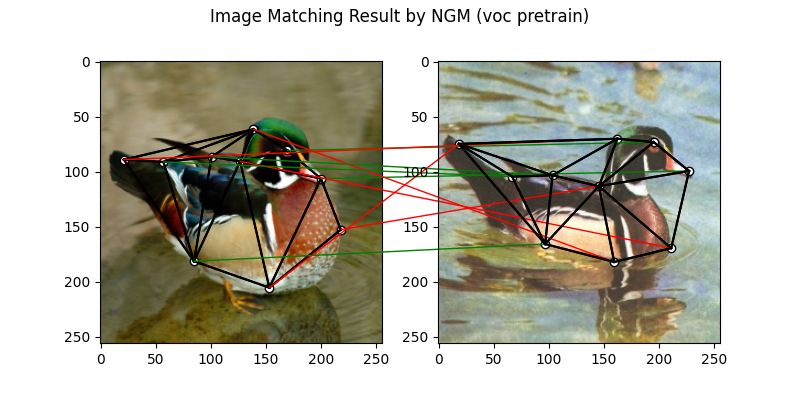
The correct matchings are marked by green, and wrong matchings are marked by red. In this example, the nodes are ordered by their ground truth classes (i.e. the ground truth matching matrix is a diagonal matrix).
plt.figure(figsize=(8, 4))
plt.suptitle('Image Matching Result by RRWM')
ax1 = plt.subplot(1, 2, 1)
plot_image_with_graph(img1, kpts1, A1)
ax2 = plt.subplot(1, 2, 2)
plot_image_with_graph(img2, kpts2, A2)
idx = jt.argmax(X, dim=1)[0]
for i in range(X.shape[0]):
j = idx[i].item()
con = ConnectionPatch(xyA=kpts1[:, i], xyB=kpts2[:, j], coordsA="data", coordsB="data",
axesA=ax1, axesB=ax2, color="red" if i != j else "green")
plt.gca().add_artist(con)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
Solve by other solvers
We could also do a quick benchmarking of other solvers on this specific problem.
IPFP solver
See ipfp() for the API reference.
X = pygm.ipfp(K, kpts1.shape[1], kpts2.shape[1])
plt.figure(figsize=(8, 4))
plt.suptitle('Image Matching Result by IPFP')
ax1 = plt.subplot(1, 2, 1)
plot_image_with_graph(img1, kpts1, A1)
ax2 = plt.subplot(1, 2, 2)
plot_image_with_graph(img2, kpts2, A2)
idx = jt.argmax(X, dim=1)[0]
for i in range(X.shape[0]):
j = idx[i].item()
con = ConnectionPatch(xyA=kpts1[:, i], xyB=kpts2[:, j], coordsA="data", coordsB="data",
axesA=ax1, axesB=ax2, color="red" if i != j else "green")
plt.gca().add_artist(con)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
SM solver
See sm() for the API reference.
X = pygm.sm(K, kpts1.shape[1], kpts2.shape[1])
X = pygm.hungarian(X)
plt.figure(figsize=(8, 4))
plt.suptitle('Image Matching Result by SM')
ax1 = plt.subplot(1, 2, 1)
plot_image_with_graph(img1, kpts1, A1)
ax2 = plt.subplot(1, 2, 2)
plot_image_with_graph(img2, kpts2, A2)
idx = jt.argmax(X, dim=1)[0]
for i in range(X.shape[0]):
j = idx[i].item()
con = ConnectionPatch(xyA=kpts1[:, i], xyB=kpts2[:, j], coordsA="data", coordsB="data",
axesA=ax1, axesB=ax2, color="red" if i != j else "green")
plt.gca().add_artist(con)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
NGM solver
See ngm() for the API reference.
Note
The NGM solvers are pretrained on a different problem setting, so their performance may seem inferior. To improve their performance, you may change the way of building affinity matrices, or try finetuning NGM on the new problem.
The NGM solver pretrained on Willow dataset:
X = pygm.ngm(K, kpts1.shape[1], kpts2.shape[1], pretrain='willow')
X = pygm.hungarian(X)
plt.figure(figsize=(8, 4))
plt.suptitle('Image Matching Result by NGM (willow pretrain)')
ax1 = plt.subplot(1, 2, 1)
plot_image_with_graph(img1, kpts1, A1)
ax2 = plt.subplot(1, 2, 2)
plot_image_with_graph(img2, kpts2, A2)
idx = jt.argmax(X, dim=1)[0]
for i in range(X.shape[0]):
j = idx[i].item()
con = ConnectionPatch(xyA=kpts1[:, i], xyB=kpts2[:, j], coordsA="data", coordsB="data",
axesA=ax1, axesB=ax2, color="red" if i != j else "green")
plt.gca().add_artist(con)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
The NGM solver pretrained on VOC dataset:
X = pygm.ngm(K, kpts1.shape[1], kpts2.shape[1], pretrain='voc')
X = pygm.hungarian(X)
plt.figure(figsize=(8, 4))
plt.suptitle('Image Matching Result by NGM (voc pretrain)')
ax1 = plt.subplot(1, 2, 1)
plot_image_with_graph(img1, kpts1, A1)
ax2 = plt.subplot(1, 2, 2)
plot_image_with_graph(img2, kpts2, A2)
idx = jt.argmax(X, dim=1)[0]
for i in range(X.shape[0]):
j = idx[i].item()
con = ConnectionPatch(xyA=kpts1[:, i], xyB=kpts2[:, j], coordsA="data", coordsB="data",
axesA=ax1, axesB=ax2, color="red" if i != j else "green")
plt.gca().add_artist(con)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: FutureWarning: The input object of type 'jittor_core.Var' is an array-like implementing one of the corresponding protocols (`__array__`, `__array_interface__` or `__array_struct__`); but not a sequence (or 0-D). In the future, this object will be coerced as if it was first converted using `np.array(obj)`. To retain the old behaviour, you have to either modify the type 'jittor_core.Var', or assign to an empty array created with `np.empty(correct_shape, dtype=object)`.
ary = asanyarray(ary)
/home/roger/.local/lib/python3.8/site-packages/numpy/core/shape_base.py:65: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
ary = asanyarray(ary)
Total running time of the script: ( 2 minutes 17.143 seconds)