Note
Go to the end to download the full example code
Jittor Backend Example: Model Fusion by Graph Matching
This example shows how to fuse different models into a single model by pygmtools.
Model fusion aims to fuse multiple models into one, such that the fused model could have higher performance.
The neural networks can be regarded as graphs (channels - nodes, update functions between channels - edges;
node feature - bias, edge feature - weights), and fusing the models is equivalent to solving a graph matching
problem. In this example, the given models are trained on MNIST data from different distributions, and the
fused model could combine the knowledge from two input models and can reach higher accuracy when testing.
# Author: Chang Liu <only-changer@sjtu.edu.cn>
# Runzhong Wang <runzhong.wang@sjtu.edu.cn>
# Wenzheng Pan <pwz1121@sjtu.edu.cn>
#
# License: Mulan PSL v2 License
Note
This is a simplified implementation of the ideas in Liu et al. Deep Neural Network Fusion via Graph Matching with Applications to Model Ensemble and Federated Learning. ICML 2022. For more details, please refer to the paper and the official code repository.
Note
The following solvers are included in this example:
sm()(classic solver)hungarian()(linear solver)
import jittor as jt
import jittor.nn as nn
from jittor import Module, transform, dataset
import time
from PIL import Image
import matplotlib.pyplot as plt
import pygmtools as pygm
pygm.set_backend('jittor')
Define a simple CNN classifier network
class SimpleNet(Module):
def __init__(self):
super(SimpleNet, self).__init__()
# self.pad = nn.ReplicationPad2d(1)
self.conv1 = nn.Conv2d(1, 32, 5, bias=False)
self.max_pool = nn.MaxPool2d(2, padding=1)
self.conv2 = nn.Conv2d(32, 64, 5, bias=False)
self.fc1 = nn.Linear(3136, 32, bias=False)
self.fc2 = nn.Linear(32, 10, bias=False)
def execute(self, x):
output = nn.relu(self.conv1(nn.ReplicationPad2d(1)(x)))
output = self.max_pool(output)
output = nn.relu(self.conv2(nn.ReplicationPad2d(1)(output)))
output = self.max_pool(output)
output = output.view(output.shape[0], -1)
output = self.fc1(output)
output = self.fc2(output)
return output
Load the trained models to be fused
model1 = SimpleNet()
model2 = SimpleNet()
model1.load_state_dict(jt.load('../data/example_model_fusion_1_jittor.dat'))
model2.load_state_dict(jt.load('../data/example_model_fusion_2_jittor.dat'))
test_transform = transform.Compose([
transform.Gray(),
transform.ToTensor()
])
test_dataset = dataset.MNIST(
data_root='../data/mnist_data_jittor/', # the directory to store the dataset
train=False, # the dataset is used to test
transform=test_transform, # the dataset is in the form of tensors
download=True)
test_loader = test_dataset.set_attrs(batch_size=32, shuffle=False)
Print the layers of the simple CNN model:
print(model1)
SimpleNet(
conv1: Conv(1, 32, (5, 5), (1, 1), (0, 0), (1, 1), 1, None, None, Kw=None, fan=None, i=None, bound=None)
max_pool: MaxPool2d(
_layer: Pool((2, 2), (2, 2), padding=(1, 1), dilation=None, return_indices=None, ceil_mode=False, count_include_pad=True, op=maximum)
)
conv2: Conv(32, 64, (5, 5), (1, 1), (0, 0), (1, 1), 1, None, None, Kw=None, fan=None, i=None, bound=None)
fc1: Linear(3136, 32, None, None)
fc2: Linear(32, 10, None, None)
)
Test the input models
with jt.no_grad():
n_correct1 = 0
n_correct2 = 0
n_samples = 0
for images, labels in test_loader:
outputs1 = model1(images)
outputs2 = model2(images)
predictions1, _ = jt.argmax(outputs1, 1)
predictions2, _ = jt.argmax(outputs2, 1)
n_samples += labels.shape[0]
n_correct1 += (predictions1 == labels).sum().item()
n_correct2 += (predictions2 == labels).sum().item()
acc1 = 100 * n_correct1 / n_samples
acc2 = 100 * n_correct2 / n_samples
Testing results (two separate models):
print(f'model1 accuracy = {acc1}%, model2 accuracy = {acc2}%')
model1 accuracy = 84.18%, model2 accuracy = 83.81%
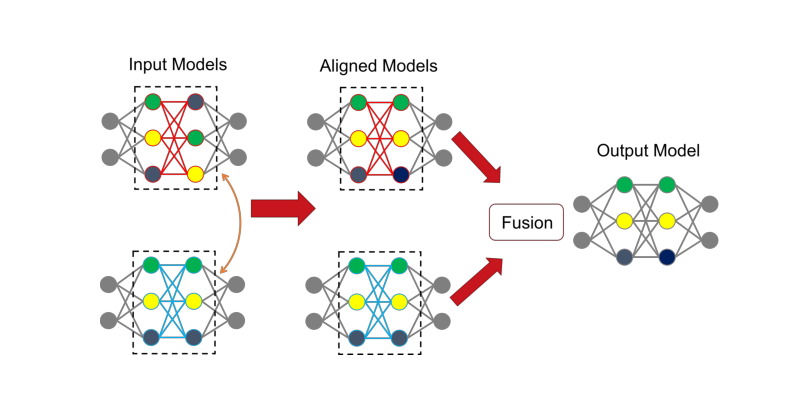
Build the affinity matrix for graph matching
As shown in the following plot, the neural networks can be regarded as graphs. The weights correspond to the edge features, and the bias corresponds to the node features. In this example, the neural network does not have bias so that there are only edge features.
plt.figure(figsize=(8, 4))
img = Image.open('../data/model_fusion.png')
plt.imshow(img)
plt.axis('off')
st_time = time.perf_counter()
Define the graph matching affinity metric function
class Ground_Metric_GM:
def __init__(self,
model_1_param: jt.Var = None,
model_2_param: jt.Var = None,
conv_param: bool = False,
bias_param: bool = False,
pre_conv_param: bool = False,
pre_conv_image_size_squared: int = None):
self.model_1_param = model_1_param
self.model_2_param = model_2_param
self.conv_param = conv_param
self.bias_param = bias_param
# bias, or fully-connected from linear
if bias_param is True or (conv_param is False and pre_conv_param is False):
self.model_1_param = self.model_1_param.reshape(1, -1, 1)
self.model_2_param = self.model_2_param.reshape(1, -1, 1)
# fully-connected from conv
elif conv_param is False and pre_conv_param is True:
self.model_1_param = self.model_1_param.reshape(1, -1, pre_conv_image_size_squared)
self.model_2_param = self.model_2_param.reshape(1, -1, pre_conv_image_size_squared)
# conv
else:
self.model_1_param = self.model_1_param.reshape(1, -1, model_1_param.shape[-1])
self.model_2_param = self.model_2_param.reshape(1, -1, model_2_param.shape[-1])
def _cdist(self, x1, x2, p = 2):
assert len(x1.shape) == len(x2.shape) == 3 and x1.shape[-1] == x2.shape[-1] # actual shape here: [1, 32, 25]
dist = jt.zeros((x1.shape[0], x1.shape[1], x2.shape[1]))
assert p == 2
for i in range(x1.shape[0]):
for j in range(x1.shape[1]):
for k in range(x2.shape[1]):
dist[i, j, k] = (x1[i][j] - x2[i][k]).pow(2).sum().sqrt().item()
return dist
def process_distance(self, p: int = 2):
return self._cdist(
self.model_1_param.float(),
self.model_2_param.float(),
p=p)[0]
def process_soft_affinity(self, p: int = 2):
return jt.exp(0 - self.process_distance(p=p))
Define the affinity function between two neural networks. This function takes multiple neural network modules, and construct the corresponding affinity matrix which is further processed by the graph matching solver.
def graph_matching_fusion(networks: list):
def total_node_num(network: jt.nn.Module):
# count the total number of nodes in the network [network]
num_nodes = 0
for idx, (name, parameters) in enumerate(network.named_parameters()):
if 'bias' in name:
continue
if idx == 0:
num_nodes += parameters.shape[1]
num_nodes += parameters.shape[0]
return num_nodes
n1 = total_node_num(network=networks[0])
n2 = total_node_num(network=networks[1])
assert (n1 == n2)
affinity = jt.zeros([n1 * n2, n1 * n2])
num_layers = len(list(zip(networks[0].parameters(), networks[1].parameters())))
num_nodes_before = 0
num_nodes_incremental = []
num_nodes_layers = []
pre_conv_list = []
cur_conv_list = []
conv_kernel_size_list = []
num_nodes_pre = 0
is_conv = False
pre_conv = False
pre_conv_out_channel = 1
is_final_bias = False
perm_is_complete = True
named_weight_list_0 = [named_parameter for named_parameter in networks[0].named_parameters()]
for idx, ((_, fc_layer0_weight), (_, fc_layer1_weight)) in \
enumerate(zip(networks[0].named_parameters(), networks[1].named_parameters())):
assert fc_layer0_weight.shape == fc_layer1_weight.shape
layer_shape = fc_layer0_weight.shape
num_nodes_cur = fc_layer0_weight.shape[0]
if len(layer_shape) > 1:
if is_conv is True and len(layer_shape) == 2:
num_nodes_pre = pre_conv_out_channel
else:
num_nodes_pre = fc_layer0_weight.shape[1]
if idx >= 1 and len(named_weight_list_0[idx - 1][1].shape) == 1:
pre_bias = True
else:
pre_bias = False
if len(layer_shape) > 2:
is_bias = False
if not pre_bias:
pre_conv = is_conv
pre_conv_list.append(pre_conv)
is_conv = True
cur_conv_list.append(is_conv)
fc_layer0_weight_data = fc_layer0_weight.data.reshape(fc_layer0_weight.shape[0], fc_layer0_weight.shape[1], -1)
fc_layer1_weight_data = fc_layer1_weight.data.reshape(fc_layer1_weight.shape[0], fc_layer1_weight.shape[1], -1)
elif len(layer_shape) == 2:
is_bias = False
if not pre_bias:
pre_conv = is_conv
pre_conv_list.append(pre_conv)
is_conv = False
cur_conv_list.append(is_conv)
fc_layer0_weight_data = fc_layer0_weight.data
fc_layer1_weight_data = fc_layer1_weight.data
else:
is_bias = True
if not pre_bias:
pre_conv = is_conv
pre_conv_list.append(pre_conv)
is_conv = False
cur_conv_list.append(is_conv)
fc_layer0_weight_data = fc_layer0_weight.data
fc_layer1_weight_data = fc_layer1_weight.data
if is_conv:
pre_conv_out_channel = num_nodes_cur
if is_bias is True and idx == num_layers - 1:
is_final_bias = True
if idx == 0:
for a in range(num_nodes_pre):
affinity[(num_nodes_before + a) * n2 + num_nodes_before + a, \
(num_nodes_before + a) * n2 + num_nodes_before + a] \
= 1
if idx == num_layers - 2 and 'bias' in named_weight_list_0[idx + 1][0] or \
idx == num_layers - 1 and 'bias' not in named_weight_list_0[idx][0]:
for a in range(num_nodes_cur):
affinity[(num_nodes_before + num_nodes_pre + a) * n2 + num_nodes_before + num_nodes_pre + a, \
(num_nodes_before + num_nodes_pre + a) * n2 + num_nodes_before + num_nodes_pre + a] \
= 1
if is_bias is False:
ground_metric = Ground_Metric_GM(
jt.Var(fc_layer0_weight_data), jt.Var(fc_layer1_weight_data), is_conv, is_bias,
pre_conv, int(fc_layer0_weight_data.shape[1] / pre_conv_out_channel))
else:
ground_metric = Ground_Metric_GM(
jt.Var(fc_layer0_weight_data), jt.Var(fc_layer1_weight_data), is_conv, is_bias,
pre_conv, 1)
layer_affinity = ground_metric.process_soft_affinity(p=2)
if is_bias is False:
pre_conv_kernel_size = fc_layer0_weight.shape[3] if is_conv else None
conv_kernel_size_list.append(pre_conv_kernel_size)
if is_bias is True and is_final_bias is False:
for a in range(num_nodes_cur):
for c in range(num_nodes_cur):
affinity[(num_nodes_before + a) * n2 + num_nodes_before + c, \
(num_nodes_before + a) * n2 + num_nodes_before + c] \
= layer_affinity[a][c]
elif is_final_bias is False:
for a in range(num_nodes_pre):
for b in range(num_nodes_cur):
affinity[
(num_nodes_before + a) * n2 + num_nodes_before:
(num_nodes_before + a) * n2 + num_nodes_before + num_nodes_pre,
(num_nodes_before + num_nodes_pre + b) * n2 + num_nodes_before + num_nodes_pre:
(num_nodes_before + num_nodes_pre + b) * n2 + num_nodes_before + num_nodes_pre + num_nodes_cur] \
= layer_affinity[a + b * num_nodes_pre].view(num_nodes_cur, num_nodes_pre).transpose(0, 1)
if is_bias is False:
num_nodes_before += num_nodes_pre
num_nodes_incremental.append(num_nodes_before)
num_nodes_layers.append(num_nodes_cur)
# affinity = (affinity + affinity.t()) / 2
return affinity, [n1, n2, num_nodes_incremental, num_nodes_layers, cur_conv_list, conv_kernel_size_list]
Get the affinity (similarity) matrix between model1 and model2.
K, params = graph_matching_fusion([model1, model2])
Align the models by graph matching
Align the channels of model1 & model2 by maximize the affinity (similarity) via graph matching algorithms.
n1 = params[0]
n2 = params[1]
X = pygm.sm(K, n1, n2)
Project X to neural network matching result. The neural network matching matrix is built by applying
Hungarian to small blocks of X, because only the channels from the same neural network layer can be
matched.
Note
In this example, we assume the last FC layer is aligned and need not be matched.
new_X = jt.zeros_like(X)
new_X[:params[2][0], :params[2][0]] = jt.init.eye(params[2][0])
for start_idx, length in zip(params[2][:-1], params[3][:-1]): # params[2] and params[3] are the indices of layers
slicing = slice(start_idx, start_idx + length)
new_X[slicing, slicing] = pygm.hungarian(X[slicing, slicing])
# assume the last FC layer is aligned
slicing = slice(params[2][-1], params[2][-1] + params[3][-1])
new_X[slicing, slicing] = jt.init.eye(params[3][-1])
X = new_X
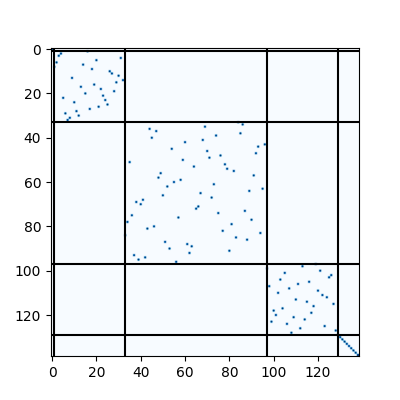
Visualization of the matching result. The black lines splits the channels of different layers.
plt.figure(figsize=(4, 4))
plt.imshow(X.numpy(), cmap='Blues')
for idx in params[2]:
plt.axvline(x=idx, color='k')
plt.axhline(y=idx, color='k')
Define the alignment function: fuse the models based on matching result
def align(solution, fusion_proportion, networks: list, params: list):
[_, _, num_nodes_incremental, num_nodes_layers, cur_conv_list, conv_kernel_size_list] = params
named_weight_list_0 = [named_parameter for named_parameter in networks[0].named_parameters()]
aligned_wt_0 = [parameter.data for name, parameter in named_weight_list_0]
idx = 0
num_layers = len(aligned_wt_0)
for num_before, num_cur, cur_conv, cur_kernel_size in \
zip(num_nodes_incremental, num_nodes_layers, cur_conv_list, conv_kernel_size_list):
perm = jt.Var(solution[num_before:num_before + num_cur, num_before:num_before + num_cur])
assert 'bias' not in named_weight_list_0[idx][0]
if len(named_weight_list_0[idx][1].shape) == 4:
aligned_wt_0[idx] = (perm.transpose(0, 1).float32() @
jt.Var(aligned_wt_0[idx]).float32().permute(2, 3, 0, 1)) \
.permute(2, 3, 0, 1)
else:
aligned_wt_0[idx] = perm.transpose(0, 1).float32() @ jt.Var(aligned_wt_0[idx]).float32()
idx += 1
if idx >= num_layers:
continue
if 'bias' in named_weight_list_0[idx][0]:
aligned_wt_0[idx] = jt.Var(aligned_wt_0[idx]).float32() @ perm.float32()
idx += 1
if idx >= num_layers:
continue
if cur_conv and len(named_weight_list_0[idx][1].shape) == 2:
aligned_wt_0[idx] = (jt.Var(aligned_wt_0[idx]).float32()
.reshape(aligned_wt_0[idx].shape[0], 64, -1)
.permute(0, 2, 1)
@ perm.float32()) \
.permute(0, 2, 1) \
.reshape(aligned_wt_0[idx].shape[0], -1)
elif len(named_weight_list_0[idx][1].shape) == 4:
aligned_wt_0[idx] = (jt.Var(aligned_wt_0[idx]).float32()
.permute(2, 3, 0, 1)
@ perm.float32()) \
.permute(2, 3, 0, 1)
else:
aligned_wt_0[idx] = jt.Var(aligned_wt_0[idx]).float32() @ perm.float32()
assert idx == num_layers
averaged_weights = []
for idx, parameter in enumerate(networks[1].parameters()):
averaged_weights.append((1 - fusion_proportion) * aligned_wt_0[idx] + fusion_proportion * parameter)
return averaged_weights
Test the fused model
The fusion_proportion variable denotes the contribution to the new model. For example, if fusion_proportion=0.2,
the fused model = 80% model1 + 20% model2.
def align_model_and_test(X):
acc_list = []
for fusion_proportion in jt.arange(0, 1.1, 0.1):
fused_weights = align(X, fusion_proportion, [model1, model2], params)
fused_model = SimpleNet()
state_dict = fused_model.state_dict()
for idx, (key, _) in enumerate(state_dict.items()):
state_dict[key] = fused_weights[idx]
fused_model.load_state_dict(state_dict)
test_loss = 0
correct = 0
for data, target in test_loader:
output = fused_model(data)
test_loss += nn.nll_loss(output, target, reduction='sum').item()
pred = output.argmax(1, keepdims=True)[0]
correct += pred.equal(target.view_as(pred)).sum()
test_loss /= test_dataset.total_len
acc = (100. * correct / test_dataset.total_len).item()
print(
f"{1 - fusion_proportion:.2f} model1 + {fusion_proportion:.2f} model2 -> fused model accuracy: {acc:.2f}%")
acc_list.append(acc)
return jt.Var(acc_list)
print('Graph Matching Fusion')
gm_acc_list = align_model_and_test(X)
Graph Matching Fusion
1.00 model1 + 0.00 model2 -> fused model accuracy: 84.18%
0.90 model1 + 0.10 model2 -> fused model accuracy: 85.12%
0.80 model1 + 0.20 model2 -> fused model accuracy: 85.21%
0.70 model1 + 0.30 model2 -> fused model accuracy: 82.52%
0.60 model1 + 0.40 model2 -> fused model accuracy: 71.11%
0.50 model1 + 0.50 model2 -> fused model accuracy: 53.74%
0.40 model1 + 0.60 model2 -> fused model accuracy: 63.26%
0.30 model1 + 0.70 model2 -> fused model accuracy: 78.51%
0.20 model1 + 0.80 model2 -> fused model accuracy: 82.81%
0.10 model1 + 0.90 model2 -> fused model accuracy: 83.97%
0.00 model1 + 1.00 model2 -> fused model accuracy: 83.81%
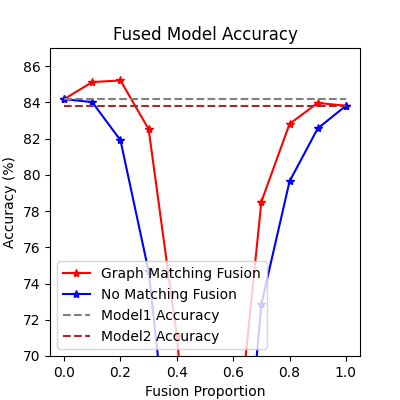
Compare with vanilla model fusion (no matching), graph matching method stabilizes the fusion step:
print('No Matching Fusion')
vanilla_acc_list = align_model_and_test(jt.init.eye(n1))
plt.figure(figsize=(4, 4))
plt.title('Fused Model Accuracy')
plt.plot(jt.arange(0, 1.1, 0.1).numpy(), gm_acc_list.numpy(), 'r*-', label='Graph Matching Fusion')
plt.plot(jt.arange(0, 1.1, 0.1).numpy(), vanilla_acc_list.numpy(), 'b*-', label='No Matching Fusion')
plt.plot(jt.arange(0, 1.1, 0.1).numpy(), [acc1] * 11, '--', color="gray", label='Model1 Accuracy')
plt.plot(jt.arange(0, 1.1, 0.1).numpy(), [acc2] * 11, '--', color="brown", label='Model2 Accuracy')
plt.gca().set_xlabel('Fusion Proportion')
plt.gca().set_ylabel('Accuracy (%)')
plt.ylim((70, 87))
plt.legend(loc=3)
plt.show()
No Matching Fusion
1.00 model1 + 0.00 model2 -> fused model accuracy: 84.18%
0.90 model1 + 0.10 model2 -> fused model accuracy: 84.01%
0.80 model1 + 0.20 model2 -> fused model accuracy: 81.91%
0.70 model1 + 0.30 model2 -> fused model accuracy: 74.67%
0.60 model1 + 0.40 model2 -> fused model accuracy: 60.39%
0.50 model1 + 0.50 model2 -> fused model accuracy: 47.16%
0.40 model1 + 0.60 model2 -> fused model accuracy: 55.34%
0.30 model1 + 0.70 model2 -> fused model accuracy: 72.86%
0.20 model1 + 0.80 model2 -> fused model accuracy: 79.64%
0.10 model1 + 0.90 model2 -> fused model accuracy: 82.56%
0.00 model1 + 1.00 model2 -> fused model accuracy: 83.81%
Print the result summary
end_time = time.perf_counter()
print(f'time consumed for model fusion: {end_time - st_time:.2f} seconds')
print(f'model1 accuracy = {acc1}%, model2 accuracy = {acc2}%')
print(f"best fused model accuracy: {jt.max(gm_acc_list):.2f}%")
time consumed for model fusion: 192.60 seconds
model1 accuracy = 84.18%, model2 accuracy = 83.81%
best fused model accuracy: 85.21%
Note
This example supports both GPU and CPU, and the online documentation is built by a CPU-only machine. The efficiency will be significantly improved if you run this code on GPU.
Total running time of the script: (3 minutes 16.258 seconds)