pygmtools.neural_solvers.cie
- pygmtools.neural_solvers.cie(feat_node1, feat_node2, A1, A2, feat_edge1, feat_edge2, n1=None, n2=None, in_node_channel=1024, in_edge_channel=1, hidden_channel=2048, out_channel=2048, num_layers=2, sk_max_iter=20, sk_tau=0.05, network=None, return_network=False, pretrain='voc', backend=None)[source]
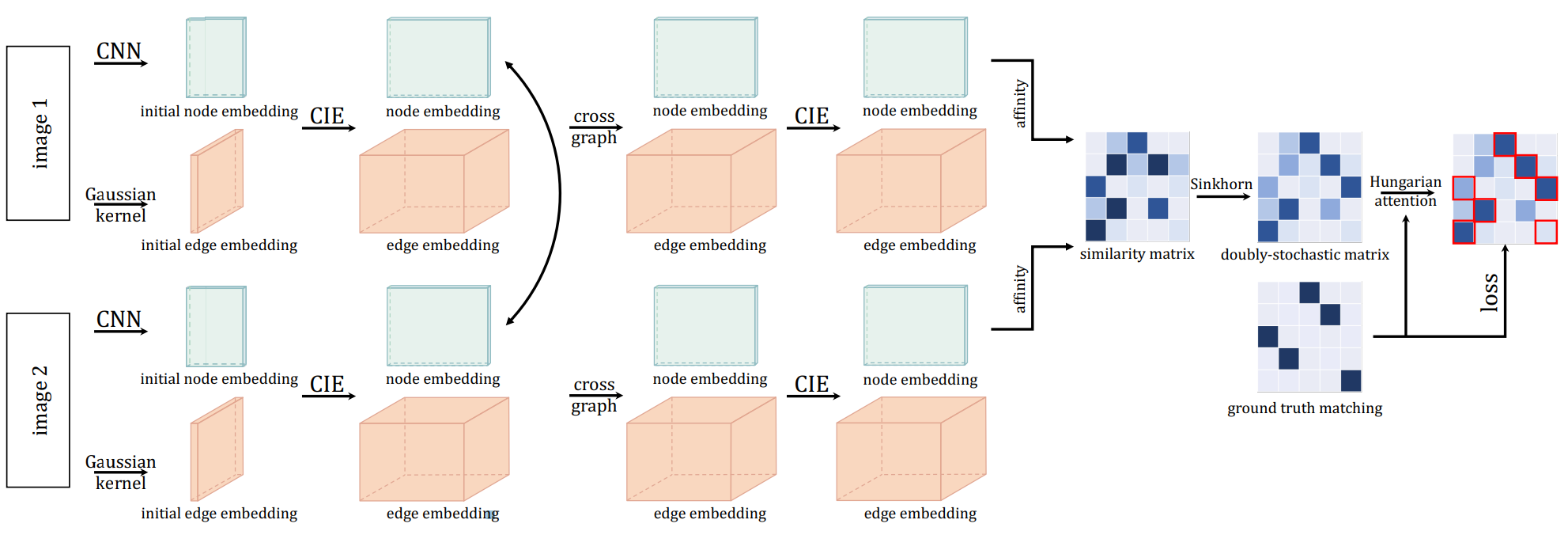
The CIE (Channel Independent Embedding) graph matching neural network model for processing two individual graphs (KB-QAP). The graph matching module is composed of several intra-graph embedding layers, a cross-graph embedding layer, and a Sinkhorn matching layer. Only the second last layer has a cross-graph update layer. The graph embedding layers are based on channel-independent embedding, under the assumption that such a message passing scheme may offer higher model capacity especially with high-dimensional edge features.
See the following pipeline for an example, with application to visual graph matching:
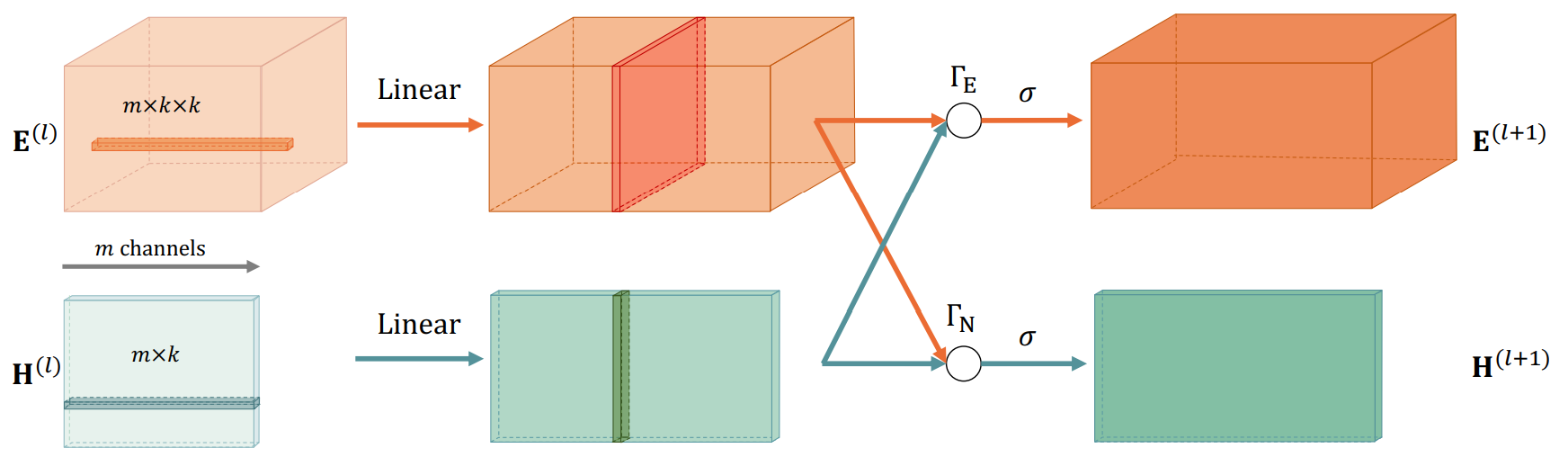
The graph embedding layer (CIE layer) involves both node embeding and edge embedding:
See the following paper for more technical details: “Yu et al. Learning Deep Graph Matching with Channel-Independent Embedding and Hungarian Attention. ICLR 2020.”
- Parameters
feat_node1 – \((b\times n_1 \times d_n)\) input node feature of graph1
feat_node2 – \((b\times n_2 \times d_n)\) input node feature of graph2
A1 – \((b\times n_1 \times n_1)\) input adjacency matrix of graph1
A2 – \((b\times n_2 \times n_2)\) input adjacency matrix of graph2
feat_edge1 – \((b\times n_1 \times n_1 \times d_e)\) input edge feature of graph1
feat_edge2 – \((b\times n_2 \times n_2 \times d_e)\) input edge feature of graph2
n1 – \((b)\) number of nodes in graph1. Optional if all equal to \(n_1\)
n2 – \((b)\) number of nodes in graph2. Optional if all equal to \(n_2\)
in_node_channel – (default: 1024) Node channel size of the input layer. It must match the feature dimension \((d_n)\) of
feat_node1, feat_node2. Ignored if the network object is given (ignored ifnetwork!=None)in_edge_channel – (default: 1) Edge channel size of the input layer. It must match the feature dimension \((d_e)\) of
feat_edge1, feat_edge2. Ignored if the network object is given (ignored ifnetwork!=None)hidden_channel – (default: 2048) Channel size of hidden layers (node channel == edge channel). Ignored if the network object is given (ignored if
network!=None)out_channel – (default: 2048) Channel size of the output layer (node channel == edge channel). Ignored if the network object is given (ignored if
network!=None)num_layers – (default: 2) Number of graph embedding layers. Must be >=2. Ignored if the network object is given (ignored if
network!=None)sk_max_iter – (default: 20) Max number of iterations of Sinkhorn. See
sinkhorn()for more details about this argument.sk_tau – (default: 0.05) The temperature parameter of Sinkhorn. See
sinkhorn()for more details about this argument.network – (default: None) The network object. If None, a new network object will be created, and load the model weights specified in
pretrainargument.return_network – (default: False) Return the network object (saving model construction time if calling the model multiple times).
pretrain – (default: ‘voc’) If
network==None, the pretrained model weights to be loaded. Available pretrained weights:voc(on Pascal VOC Keypoint dataset),willow(on Willow Object Class dataset), orFalse(no pretraining).backend – (default:
pygmtools.BACKENDvariable) the backend for computation.
- Returns
if
return_network==False, \((b\times n_1 \times n_2)\) the doubly-stochastic matching matrixif
return_network==True, \((b\times n_1 \times n_2)\) the doubly-stochastic matching matrix, the network object
Note
You may need a proxy to load the pretrained weights if Google drive is not accessible in your contry/region. You may also download the pretrained models manually and put them at
~/.cache/pygmtools(for Linux).Note
This function also supports non-batched input, by ignoring all batch dimensions in the input tensors.
Numpy Example
>>> import numpy as np >>> import pygmtools as pygm >>> pygm.set_backend('numpy') >>> np.random.seed(1) # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> X_gt = np.zeros((batch_size, 4, 4)) >>> X_gt[:, np.arange(0, 4, dtype='i4'), np.random.permutation(4)] = 1 >>> A1 = 1. * (np.random.rand(batch_size, 4, 4) > 0.5) >>> for i in np.arange(4): # discard self-loop edges ... for j in np.arange(batch_size): ... A1[j][i][i] = 0 >>> e_feat1 = np.expand_dims(np.random.rand(batch_size, 4, 4) * A1,axis=-1) # shape: (10, 4, 4, 1) >>> A2 = np.matmul(np.matmul(X_gt.swapaxes(1, 2), A1), X_gt) >>> e_feat2 = np.expand_dims(np.matmul(np.matmul(X_gt.swapaxes(1, 2),np.squeeze(e_feat1,axis=-1)), X_gt),axis=-1) >>> feat1 = np.random.rand(batch_size, 4, 1024) - 0.5 >>> feat2 = np.matmul(X_gt.swapaxes(1, 2), feat1) >>> n1 = n2 = np.array([4] * batch_size) # Match by CIE (load pretrained model) >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True) Downloading to ~/.cache/pygmtools/cie_voc_numpy.npy... >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy 1.0 # Pass the net object to avoid rebuilding the model agian >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy 1.0 # You may also load other pretrained weights >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True, pretrain='willow') Downloading to ~/.cache/pygmtools/cie_willow_numpy.npy... >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy 1.0 # You may configure your own model and integrate the model into a deep learning pipeline. For example: >>> net = pygm.utils.get_network(pygm.cie, in_node_channel=1024, in_edge_channel=1, hidden_channel=2048, out_channel=512, num_layers=3, pretrain=False) # feat1/feat2/e_feat1/e_feat2 may be outputs by other neural networks >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy 1.0
PyTorch Example
>>> import torch >>> import pygmtools as pygm >>> pygm.set_backend('pytorch') >>> _ = torch.manual_seed(1) # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> X_gt = torch.zeros(batch_size, 4, 4) >>> X_gt[:, torch.arange(0, 4, dtype=torch.int64), torch.randperm(4)] = 1 >>> A1 = 1. * (torch.rand(batch_size, 4, 4) > 0.5) >>> torch.diagonal(A1, dim1=1, dim2=2)[:] = 0 # discard self-loop edges >>> e_feat1 = (torch.rand(batch_size, 4, 4) * A1).unsqueeze(-1) # shape: (10, 4, 4, 1) >>> A2 = torch.bmm(torch.bmm(X_gt.transpose(1, 2), A1), X_gt) >>> e_feat2 = torch.bmm(torch.bmm(X_gt.transpose(1, 2), e_feat1.squeeze(-1)), X_gt).unsqueeze(-1) >>> feat1 = torch.rand(batch_size, 4, 1024) - 0.5 >>> feat2 = torch.bmm(X_gt.transpose(1, 2), feat1) >>> n1 = n2 = torch.tensor([4] * batch_size) # Match by CIE (load pretrained model) >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True) Downloading to ~/.cache/pygmtools/cie_voc_pytorch.pt... >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy tensor(1.) # Pass the net object to avoid rebuilding the model agian >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) # You may also load other pretrained weights >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True, pretrain='willow') Downloading to ~/.cache/pygmtools/cie_willow_pytorch.pt... # You may configure your own model and integrate the model into a deep learning pipeline. For example: >>> net = pygm.utils.get_network(pygm.cie, in_node_channel=1024, in_edge_channel=1, hidden_channel=2048, out_channel=512, num_layers=3, pretrain=False) >>> optimizer = torch.optim.SGD(net.parameters(), lr=0.001, momentum=0.9) # feat1/feat2/e_feat1/e_feat2 may be outputs by other neural networks >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) >>> loss = pygm.utils.permutation_loss(X, X_gt) >>> loss.backward() >>> optimizer.step()
Jittor Example
>>> import jittor as jt >>> import pygmtools as pygm >>> pygm.set_backend('jittor') >>> _ = jt.seed(1) # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> X_gt = jt.zeros((batch_size, 4, 4)) >>> X_gt[:, jt.arange(0, 4, dtype=jt.int64), jt.randperm(4)] = 1 >>> A1 = 1. * (jt.rand(batch_size, 4, 4) > 0.5) >>> for i in range(batch_size): >>> for j in range(4): >>> A1.data[i][j][j] = 0 # discard self-loop edges >>> e_feat1 = (jt.rand(batch_size, 4, 4) * A1).unsqueeze(-1) # shape: (10, 4, 4, 1) >>> A2 = jt.bmm(jt.bmm(X_gt.transpose(1, 2), A1), X_gt) >>> e_feat2 = jt.bmm(jt.bmm(X_gt.transpose(1, 2), e_feat1.squeeze(-1)), X_gt).unsqueeze(-1) >>> feat1 = jt.rand(batch_size, 4, 1024) - 0.5 >>> feat2 = jt.bmm(X_gt.transpose(1, 2), feat1) >>> n1 = n2 = jt.Var([4] * batch_size) # Match by CIE (load pretrained model) >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True) Downloading to ~/.cache/pygmtools/cie_voc_jittor.pt... >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy jt.Var([1.], dtype=float32) # Pass the net object to avoid rebuilding the model agian >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) # You may also load other pretrained weights >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True, pretrain='willow') Downloading to ~/.cache/pygmtools/cie_willow_jittor.pt... # You may configure your own model and integrate the model into a deep learning pipeline. For example: >>> net = pygm.utils.get_network(pygm.cie, in_node_channel=1024, in_edge_channel=1, hidden_channel=2048, out_channel=512, num_layers=3, pretrain=False) >>> optimizer = jt.optim.SGD(net.parameters(), lr=0.001, momentum=0.9) # feat1/feat2/e_feat1/e_feat2 may be outputs by other neural networks >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) >>> loss = pygm.utils.permutation_loss(X, X_gt) >>> optimizer.backward(loss) >>> optimizer.step()
Paddle Example
>>> import paddle >>> import pygmtools as pygm >>> pygm.set_backend('paddle') >>> _ = paddle.seed(1) # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> X_gt = paddle.zeros((batch_size, 4, 4)) >>> X_gt[:, paddle.arange(0, 4, dtype=paddle.int64), paddle.randperm(4)] = 1 >>> A1 = 1. * (paddle.rand((batch_size, 4, 4)) > 0.5) >>> paddle.diagonal(A1, axis1=1, axis2=2)[:] = 0 # discard self-loop edges >>> e_feat1 = (paddle.rand((batch_size, 4, 4)) * A1).unsqueeze(-1) # shape: (10, 4, 4, 1) >>> A2 = paddle.bmm(paddle.bmm(X_gt.transpose((0, 2, 1)), A1), X_gt) >>> e_feat2 = paddle.bmm(paddle.bmm(X_gt.transpose((0, 2, 1)), e_feat1.squeeze(-1)), X_gt).unsqueeze(-1) >>> feat1 = paddle.rand((batch_size, 4, 1024)) - 0.5 >>> feat2 = paddle.bmm(X_gt.transpose((0, 2, 1)), feat1) >>> n1 = n2 = paddle.to_tensor([4] * batch_size) # Match by CIE (load pretrained model) >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True) Downloading to ~/.cache/pygmtools/cie_voc_paddle.pdparams... >>> (pygm.hungarian(X) * X_gt).sum() / X_gt.sum() # accuracy Tensor(shape=[1], dtype=float32, place=Place(cpu), stop_gradient=True, [1.]) # Pass the net object to avoid rebuilding the model agian >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) # You may also load other pretrained weights >>> X, net = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, return_network=True, pretrain='willow') Downloading to ~/.cache/pygmtools/cie_willow_paddle.pdparams... # You may configure your own model and integrate the model into a deep learning pipeline. For example: >>> net = pygm.utils.get_network(pygm.cie, in_node_channel=1024, in_edge_channel=1, hidden_channel=2048, out_channel=512, num_layers=3, pretrain=False) >>> optimizer = paddle.optimizer.SGD(parameters=net.parameters(), learning_rate=0.001) # feat1/feat2/e_feat1/e_feat2 may be outputs by other neural networks >>> X = pygm.cie(feat1, feat2, A1, A2, e_feat1, e_feat2, n1, n2, network=net) >>> loss = pygm.utils.permutation_loss(X, X_gt) >>> loss.backward() >>> optimizer.step()
Note
If you find this model useful in your research, please cite:
@inproceedings{YuICLR20, title={Learning deep graph matching with channel-independent embedding and Hungarian attention}, author={Yu, Tianshu and Wang, Runzhong and Yan, Junchi and Li, Baoxin}, booktitle={International Conference on Learning Representations}, year={2020} }