pygmtools.neural_solvers.genn_astar
- pygmtools.neural_solvers.genn_astar(feat1, feat2, A1, A2, n1=None, n2=None, channel=None, filters_1=64, filters_2=32, filters_3=16, tensor_neurons=16, beam_width=0, trust_fact=1, no_pred_size=0, network=None, return_network=False, pretrain='AIDS700nef', backend=None)[source]
The GENN-A* (Graph Edit Neural Network A*) solver for graph matching (and graph edit distance) based on the fusion of traditional A* and Neural Network. This algorithm replaces the heuristic prediction module in the traditional A* algorithm with GENN (Graph Edit Neural Network) model, greatly improving the efficiency of A* algorithm with negligible loss in accuracy. During the search process, the algorithm chooses the next state with the lowest edit cost, which is predicted by GENN.
See the following picture to better understand the workflow of the algorithm:
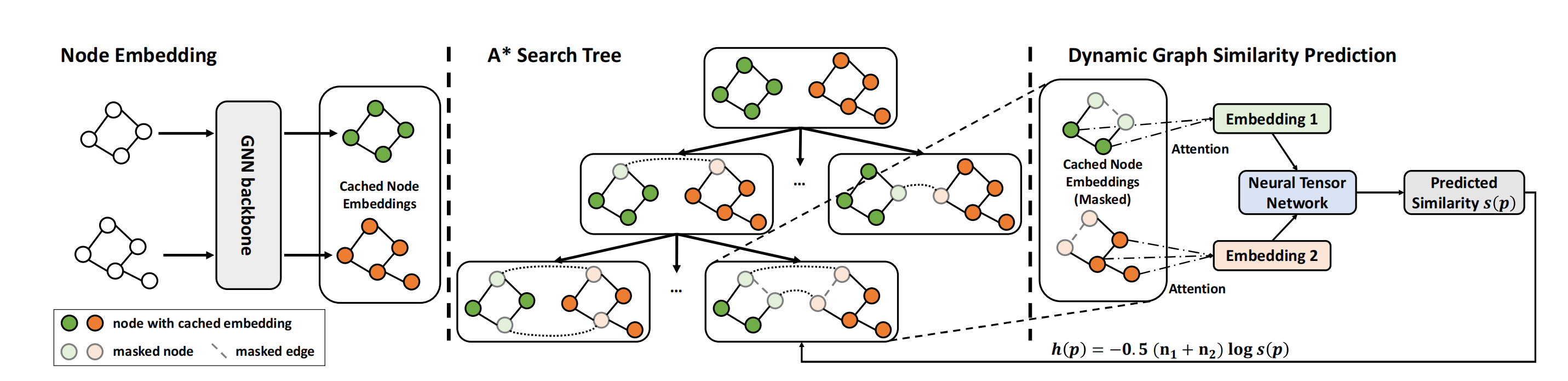
See the following paper for more technical details: “Combinatorial Learning of Graph Edit Distance via Dynamic Embedding”
- Parameters
feat1 – \((b\times n_1 \times d)\) input feature of graph1
feat2 – \((b\times n_2 \times d)\) input feature of graph2
A1 – \((b\times n_1 \times n_1)\) input adjacency matrix of graph1
A2 – \((b\times n_2 \times n_2)\) input adjacency matrix of graph2
n1 – \((b)\) number of nodes in graph1. Optional if all equal to \(n_1\)
n2 – \((b)\) number of nodes in graph2. Optional if all equal to \(n_2\)
channel – (default: None) Channel size of the input layer. If given, it must match the feature dimension (d) of feat1, feat2. If not given, it will be defined by the feature dimension (d) of feat1, feat2. Ignored if the network object isgiven (ignored if network!=None)
filters_1 – (default: 64) Filters (neurons) in 1st convolution.
filters_2 – (default: 32) Filters (neurons) in 2nd convolution.
filters_3 – (default: 16) Filters (neurons) in 2nd convolution.
tensor_neurons – (default: 16) Neurons in tensor network layer.
beam_width – (default: 0) Size of beam-search witdh (0 = no beam).
trust_fact – (default: 1) The trust factor on GNN prediction (0 = no GNN).
no_pred_size – (default: 0) If the smaller graph has no more than
no_pred_sizenodes, stop using heuristics.network – (default: None) The network object. If None, a new network object will be created, and load the model weights specified in
pretrainargument.return_network – (default: False) Return the network object (saving model construction time if calling the model multiple times).
pretrain – (default: ‘AIDS700nef’) If
network==None, the pretrained model weights to be loaded. Available pretrained weights:AIDS700nef(channel=36),LINUX(channel=8), orFalse(no pretraining).backend – (default:
pygmtools.BACKENDvariable) the backend for computation.
- Returns
if
return_network==False, \((b\times n_1 \times n_2)\) the doubly-stochastic matching matrixif
return_network==True, \((b\times n_1 \times n_2)\) the doubly-stochastic matching matrix, the network object
Note
Graph matching problem (Lawler’s QAP) and graph edit distance problem are two sides of the same coin. If you want to transform your graph edit distance problem into Lawler’s QAP, first compute the edit cost of each node pairs and edge pairs, then place them to the right places to get a cost matrix. Finally, build the affinity matrix by \(-1\times\) cost matrix.
You can get your customized cost/affinity matrix by
build_aff_mat().Note
You may need a proxy to load the pretrained weights if Google drive is not accessible in your contry/region. You may also download the pretrained models manually and put them at
~/.cache/pygmtools(for Linux).Note
This function also supports non-batched input, by ignoring all batch dimensions in the input tensors.
PyTorch Example
>>> import torch >>> import pygmtools as pygm >>> pygm.set_backend('pytorch') >>> _ = torch.manual_seed(1) # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> nodes_num = 4 >>> channel = 36 >>> X_gt = torch.zeros(batch_size, nodes_num, nodes_num) >>> X_gt[:, torch.arange(0, nodes_num, dtype=torch.int64), torch.randperm(nodes_num)] = 1 >>> A1 = 1. * (torch.rand(batch_size, nodes_num, nodes_num) > 0.5) >>> torch.diagonal(A1, dim1=1, dim2=2)[:] = 0 # discard self-loop edges >>> A2 = torch.bmm(torch.bmm(X_gt.transpose(1, 2), A1), X_gt) >>> feat1 = torch.rand(batch_size, nodes_num, channel) - 0.5 >>> feat2 = torch.bmm(X_gt.transpose(1, 2), feat1) >>> n1 = n2 = torch.tensor([nodes_num] * batch_size) # Match by GENN-A* (load pretrained model) >>> X, net = pygm.genn_astar(feat1, feat2, A1, A2, n1, n2, return_network=True) Downloading to ~/.cache/pygmtools/best_genn_AIDS700nef_gcn_astar.pt... >>> (X * X_gt).sum() / X_gt.sum()# accuracy tensor(1.) # Pass the net object to avoid rebuilding the model agian >>> X = pygm.genn_astar(feat1, feat2, A1, A2, n1, n2, network=net) # This function also supports non-batched input, by ignoring all batch dimensions in the input tensors. >>> part_f1 = feat1[0] >>> part_f2 = feat2[0] >>> part_A1 = A1[0] >>> part_A2 = A2[0] >>> part_X_gt = X_gt[0] >>> part_X = pygm.genn_astar(part_f1, part_f2, part_A1, part_A2, return_network=False) >>> part_X.shape torch.Size([4, 4]) >>> (part_X * part_X_gt).sum() / part_X_gt.sum()# accuracy tensor(1.) # You may also load other pretrained weights # However, it should be noted that each pretrained set supports different node feature dimensions # AIDS700nef(Default): channel = 36 # LINUX: channel = 8 # Generate a batch of isomorphic graphs >>> batch_size = 10 >>> nodes_num = 4 >>> channel = 8 >>> X_gt = torch.zeros(batch_size, nodes_num, nodes_num) >>> X_gt[:, torch.arange(0, nodes_num, dtype=torch.int64), torch.randperm(nodes_num)] = 1 >>> A1 = 1. * (torch.rand(batch_size, nodes_num, nodes_num) > 0.5) >>> torch.diagonal(A1, dim1=1, dim2=2)[:] = 0 # discard self-loop edges >>> A2 = torch.bmm(torch.bmm(X_gt.transpose(1, 2), A1), X_gt) >>> feat1 = torch.rand(batch_size, nodes_num, channel) - 0.5 >>> feat2 = torch.bmm(X_gt.transpose(1, 2), feat1) >>> n1 = n2 = torch.tensor([nodes_num] * batch_size) >>> X, net = pygm.genn_astar(feat1, feat2, A1, A2, n1, n2, pretrain='LINUX', return_network=True) Downloading to ~/.cache/pygmtools/best_genn_LINUX_gcn_astar.pt... >>> (X * X_gt).sum() / X_gt.sum()# accuracy tensor(1.) # When the input node feature dimension is different from the one supported by pre training, # you can still use the solver, but the solver will provide a warning >>> X, net = pygm.genn_astar(feat1, feat2, A1, A2, n1, n2, return_network=True, pretrain='AIDS700nef') Warning: Skip key(s) in state_dict: "convolution_1.weight". Warning: Pretrain AIDS700nef does not support the parameters you entered, Supported parameters: ( channel:36, filters:(64,32,16) ), Input parameters: ( channel:8, filters:(64,32,16) ) # You may configure your own model and integrate the model into a deep learning pipeline. For example: >>> net = pygm.utils.get_network(pygm.genn_astar, channel = 1000, filters_1 = 1024, filters_2 = 256, filters_3 = 128, pretrain=False) >>> optimizer = torch.optim.SGD(net.parameters(), lr=0.001, momentum=0.9) # feat1/feat2 may be outputs by other neural networks >>> X = pygm.genn_astar(feat1, feat2, A1, A2, n1, n2, network=net) >>> loss = pygm.utils.permutation_loss(X, X_gt) >>> loss.backward() >>> optimizer.step()
Note
If you find this model useful in your research, please cite:
@inproceedings{WangCVPR21, author={Runzhong Wang, Tianqi Zhang, Tianshu Yu, Junchi Yan, Xiaokang Yang}, booktitle={2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)}, title={Combinatorial Learning of Graph Edit Distance via Dynamic Embedding}, year={2021}, }